anavar_utils
Introduction
anavar_utils is a python package designed to facilitate the easy creation of control files for anavar (Barton and Zeng, 2018) as well as providing classes for the easy interpretation of anavar’s results files.
Anavar is available here, the manual describes in much more detail the models available and the parameters in the control files than in these docs.
To use the package to create a simple control file:
from __future__ import print_function
import anavar_utils as an
# data needed for anavar, SFS, callable sites and sample size
sample_size = 10
site_frequencies = [20, 19, 18, 17, 16, 15, 2, 1]
n_sites = 10000
# anavar_utils takes the sfs data in a dictionary
sfs_dict = {'SNP': (site_frequencies, n_sites)}
# intiate control file instance
control_file = an.Snp1ControlFile()
# set the data
control_file.set_data(sfs_dict, sample_size)
# construct control file string
control_contents = control_file.construct()
# output to a file or to stdout
print(control_contents)
This gives:
[algorithm_commands]
search_algorithm: NLOPT_LD_LBFGS
maxeval: 100000
maxtime: 600
num_searches: 500
nnoimp: 1
maximp: 3
optional: false
[model_commands]
model: SNP_1
n: 17
m: 10000
folded: false
sfs: 20, 19, 18, 17, 16, 15, 2, 1
dfe: discrete
c: 1
theta_range: 1e-06, 0.1
gamma_range: -250, 10
e_range: 0.0, 0.5
constraint: none
API
Control Files
Site frequency data format
In its simplest form a control file can be created for a given model
with only the site frequency spectrum, the number of callable sites
(in some cases and if per site parameter estimates are not required
this can be set to 0) and the sample size. The control file classes
take this data (using the .set_data() method) in the form of a
dictionary, with keys specific to each model, as show in the table
below.
| model | dictionary format |
|---|---|
| SNP_1 | {'SNP': (sfs, n_sites)} |
| INDEL_1 | {'INS': (sfs, n_sites), 'DEL': (sfs, n_sites)} |
| gBGC_GLEMIN_EXTENDED_M1* | {'neutral_SNPs': (sfs, n_sites), 'ws_SNPs': (sfs, n_sites), 'sw_SNPs': (sfs, n_sites)} |
| neutralINDEL_vs_selectedINDEL | {'neutral_INS': (sfs, n_sites), 'neutral_DEL': (sfs, n_sites), 'selected_INS': (sfs, n_sites), 'selected_DEL': (sfs, n_sites)} |
| neutralSNP_vs_selectedSNP | {'neutral_SNP': (sfs, n_sites), 'selected_SNP': (sfs, n_sites)} |
The site frequency data needs to provide in the form of frequency counts from low freq to high freq with frequencies with no variants entered as 0. For example this site frequency spectrum (SFS):
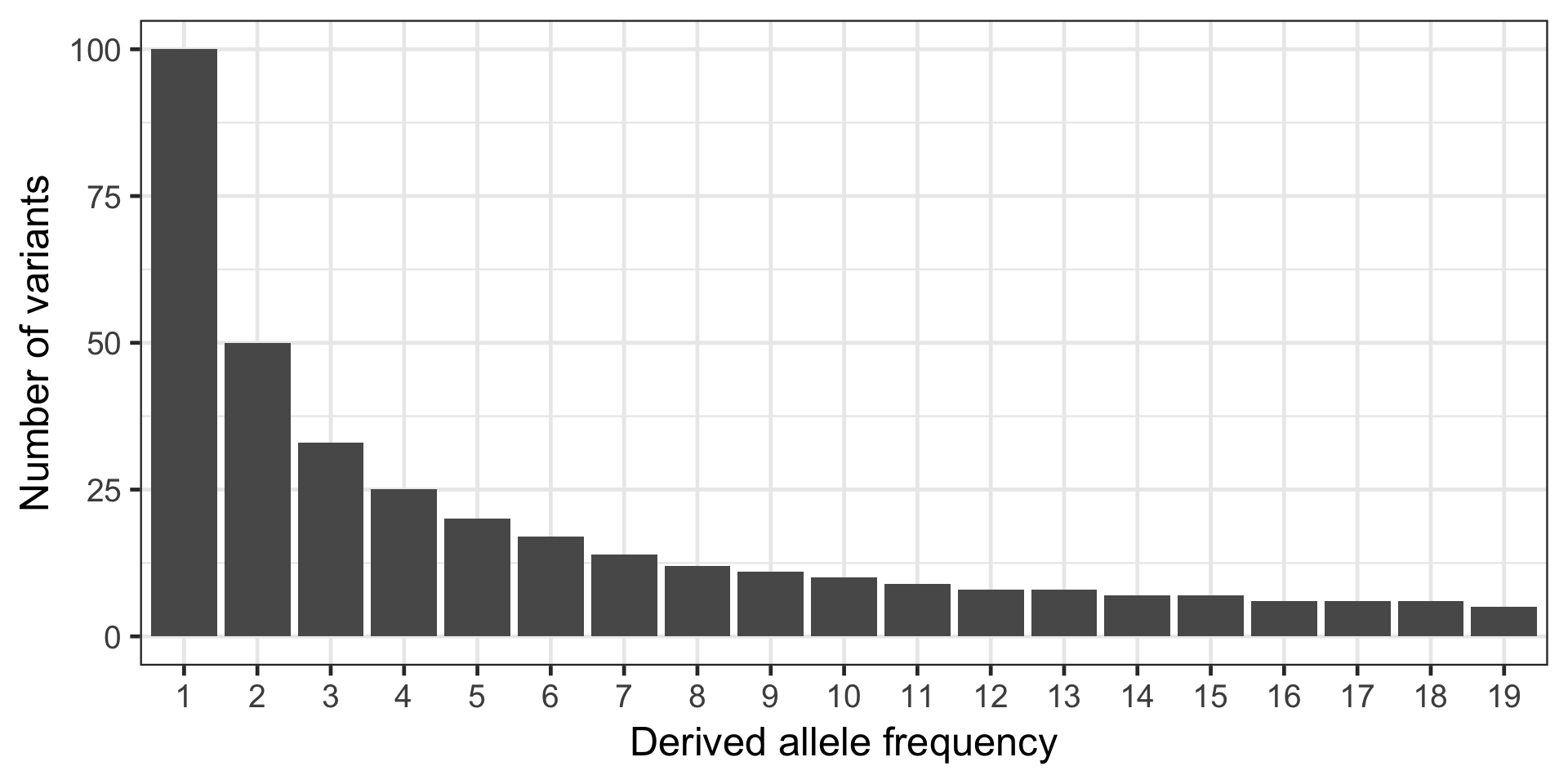
Would be expressed as:
sfs = [100, 50, 33, 25, 20, 17, 14, 12, 11, 10, 9, 8, 8, 7, 7, 6, 6, 6, 5]
class Snp1ControlFile
Snp1ControlFile()
Initiates instance of class to be used for creating a control file for the SNP_1 model. Also the parent class for all other control file classes.
construct(self)
Creates the final control file string with all specified settings
set_alg_opts(self, alg='NLOPT_LD_LBFGS', maxeval=100000, maxtime=600, search=500, nnoimp=1, maximp=3, optional=False, size=10000, key=3, epsabs=1e-50, epsrel=1e-10, rftol=1e-10, init=())
Sets algorithm options in control file
Parameters:
-
alg - (str) - default’NLOPT_LD_LBFGS’: the algorithm to use, choose from: ‘NLOPT_LD_SLSQP’, ‘NLOPT_LD_LBFGS’, ‘NLOPT_LD_VAR1’, ‘NLOPT_LD_VAR2’, ‘NLOPT_LN_NELDERMEAD’ or ‘NLOPT_LD_TNEWTON_PRECOND_RESTART’
-
maxeval - (int) - default=100000: search will be aborted when the likelihood function has been evaluated this number of times but the algorithm is still deemed non-converged
-
maxtime - (int) - default=600: search will be aborted when the algorithm has been running for the given amount of time (in seconds)
-
search - (int) - default=500: number of independent searches to be carried out
-
nnoimp - (int) - default=1: if is set to 1, then no improvement search is attempted, otherwise, improvement searches will be conducted until nnoimp number of consecutive improvement attempts lead to no significant improvement in the ln-likelihood
-
maximp - (int) - default=3: maximum number of improvement searches
-
optional - (bool) - default=False: if True, then optional commands will be included (optional commands are set separately, but will not be included until optional is set to True)
-
size - (int) - default=10000: controls QAG adaptive integration algorithm in GSL see: https://www.gnu.org/software/gsl/manual/html_node/QAG-adaptive-integration.html
-
key - (int) - default=3: controls QAG adaptive integration algorithm in GSL see: https://www.gnu.org/software/gsl/manual/html_node/QAG-adaptive-integration.html
-
epsabs - (float) - default (anavar_utils)=1e-50 - default (anavar1.4)=1e-5: controls QAG adaptive integration algorithm in GSL see: https://www.gnu.org/software/gsl/manual/html_node/QAG-adaptive-integration.html
-
epsrel - (float) - default (anavar_utils)=1e-10 - default (anavar1.4)=1e-8: controls QAG adaptive integration algorithm in GSL see: https://www.gnu.org/software/gsl/manual/html_node/QAG-adaptive-integration.html
-
rftol - (float) - default (anavar_utils)=1e-10 - default (anavar1.4)=1e-8: when consecutive search steps differ by less than the given value then the algorithm is considered to have converged
-
init - (tuple) - default=(): takes a tuple of starting values of all parameters (in order of appearance in results file) for the model, default gives random starting values within parameter ranges
set_constraint(self, constraint)
Sets model constraints in control file
Parameters:
- constraint - (str) - default=’none’: sets model constraint, chose form: ‘none’, ‘no_pol_error’, ‘equal_pol_error’, ‘equal_mutation_rate’
set_data(self, sfs_m, n, snp_fold=False, dfe='discrete', c=1, theta_r=(1e-06, 0.1), gamma_r=(-250, 10), error_r=(0.0, 0.5), shape_r=(0.001, 200), scale_r=(0.1, 1000.0), r_r=(0.05, 5.0))
Sets model and dfe commands in control file
Parameters:
-
sfs_m - (dict) - required: site frequency data in the form of {key: (sfs_list, number_call_sites)} see table at beginning of API section for the relevant dictionary structure for each model
-
n - (int) - required: sample size
-
snp_fold - (bool) - default=False: if specified sets folded to True, i.e. informs the model that a folded (unpolarised) SFS is given
-
dfe - (str) - default=’discrete’: sets the type of DFE to use, choose from ‘discrete’ or ‘continuous’
-
c - (int) - default=1: if dfe=’discrete’ then c determines the number of classes of sites to estimate parameters for
-
theta_r - (tuple) - default=(1e-06, 0.1): specifies the lower and upper theta limits in the form (lower, upper)
-
gamma_r - (tuple) - default=(-250, 10): specifies the lower and upper gamma limits in the form (lower, upper)
-
error_r - (tuple) - default=(0.0, 0.5): specifies the lower and upper polarisation error limits in the form (lower, upper)
-
shape_r - (tuple) - default=(0.001, 200): specifies the lower and upper polarisation shape parameter limits in the form (lower, upper)
-
scale_r - (tuple) - default=(0.1, 1000.0): specifies the lower and upper polarisation scale parameter limits in the form (lower, upper)
-
r_r - (tuple) - default=(0.05, 5.0): specifies the lower and upper polarisation r parameter limits in the form (lower, upper)
set_dfe_optional_opts(self, optional=False, fraction=0.005, degree=50, delta=1e-05)
Sets optional commands for the DFE
Parameters:
-
optional - (bool) - default=False: if True optional commands will be added to control file
-
fraction - (float) - default=0.005: for controlling the step size, h, used in numerical differentiation
-
degree - (int) - default=50: the integration over the Gamma distribution is approximated by an n-degree Gaussian quadrature
-
delta - (float) - default=1e-5: for controlling the step size, h, used in numerical differentiation
class GbgcControlFile
GbgcControlFile(Snp1ControlFile)
Initiates instance of class to be used for creating a control file for the gBGC_EXTENDED_M1* model.
Inherits methods from Snp1ControlFile.
set_constraint(self, constraint)
Sets model constraints in control file
Parameters:
- constraint - (str) - default=’none’: sets model constraint, chose form: ‘none’, ‘M0’, ‘M1’, ‘M0’ and ‘M1’
class Indel1ControlFile
Indel1ControlFile(Snp1ControlFile)
Initiates instance of class to be used for creating a control file for the INDEL_1 model.
Inherits methods from Snp1ControlFile.
set_constraint(self, constraint)
Sets model constraints in control file
Parameters:
- constraint - (str) - default=’none’: sets model constraint, chose form: ‘none’ and ‘neutral’
class IndelNeuSelControlFile
IndelNeuSelControlFile(Snp1ControlFile)
Initiates instance of class to be used for creating a control file for the neutralINDEL_vs_selectedINDEL model.
Inherits methods from Snp1ControlFile.
set_constraint(self, constraint)
Sets model constraints in control file
Parameters:
- constraint - (str) - default=’none’: sets model constraint, chose form: ‘none’ and ‘equal_mutation_rate’
class SNPNeuSelControlFile
SNPNeuSelControlFile(Snp1ControlFile)
Initiates instance of class to be used for creating a control file for the neutralSNP_vs_selectedSNP model.
Inherits methods from Snp1ControlFile.
set_constraint(self, constraint)
Sets model constraints in control file
Parameters:
- constraint - (str) - default=’none’: sets model constraint, chose form: ‘none’ and ‘equal_mutation_rate’
Results Files
class ResultsFile
ResultsFile(self, anavar_results_file)
Creates an anavar results file object from a file object of a valid anavar results file
Parameters:
- anavar_results_file - (file_object) - required: an open anavar results file
bounds_hit(self, theta_r=(1e-06, 0.1), gamma_r=(-250, 10), error_r=(0.0, 0.5), shape_r=(0.001, 200), scale_r=(0.1, 1000.0), r_r=(0.05, 5.0))
Determines if any of the parameter estimates in the results file have hit their upper or lower limits
Parameters:
-
theta_r - (tuple) - default=(1e-06, 0.1): specifies the lower and upper theta limits in the form (lower, upper)
-
gamma_r - (tuple) - default=(-250, 10): specifies the lower and upper gamma limits in the form (lower, upper)
-
error_r - (tuple) - default=(0.0, 0.5): specifies the lower and upper polarisation error limits in the form (lower, upper)
-
shape_r - (tuple) - default=(0.001, 200): specifies the lower and upper polarisation shape parameter limits in the form (lower, upper)
-
scale_r - (tuple) - default=(0.1, 1000.0): specifies the lower and upper polarisation scale parameter limits in the form (lower, upper)
-
r_r - (tuple) - default=(0.05, 5.0): specifies the lower and upper polarisation r parameter limits in the form (lower, upper)
Returns:
- list - parameters that hit limits
control_file(self)
Returns:
- str - control file path listed in results file
converged(self)
Determines if algorithm has converged. If ln likelihoods differ by less than 0.1 algorithm is said to have converged
Returns:
- bool - convergence status results file
data_type(self)
Returns:
- str - returns either ‘indel’, ‘snp’ or ‘snp_indel’
dfe(self)
Returns:
- str - returns distribution type ‘continuous’ or ‘discrete’
estimates(self, as_string=False)
Parameters:
- as_string - bool - default=False: if True will yield each results line as a string, if False yields each line as a dictionary with the parameter names as keys
Yields:
- str or dict - yields each results line of the file
free_parameters(self)
Returns:
- tuple - the free parameters in the model, as found at the top of the results file
get_alpha(self, dn, ds, var_type)
Calculates alpha (proportion of substitutions fixed by positive selection) using equation 19 in Barton and Zeng (2018)
Parameters:
-
dn - (float) - required: non-synonymous divergence
-
ds - (float) - required: synonymous divergence
-
var_type - (str) - required: variant type to calculate alpha for, choose from ‘snp’, ‘indel’, ‘ins’, and ‘del’
Returns:
- float - alpha value
header(self)
Returns:
- tuple - returns the column headers line from the file
ml_estimate(self, as_string=False)
Gets the maximum likelihood estimate from the results file (assumes a sorted results file)
Parameters:
- as_string - bool - default=False: if True will returns line as a string, if False returns line as a dictionary with the parameter names as keys
Returns:
- str or dict - returns maximum likelihood parameter estimates
num_class(self)
Returns:
- int - returns number of classes of sites in results (c), if dfe=’continuous’ returns 1
num_runs(self)
Returns:
- int - gives the number of runs listed in the results file
seed(self)
Returns:
- int - returns the seed anavar was run with
class MultiResultsFile
MultiResultsFile(self, file_list)
Merges anavar results files by creating ResultsFile instances for each file and writing a temporary results file of merged results before calling a ResultsFile instance on the new merged file, which is then deleted.
Parameters:
- file_list - (list) - required: list of anavar results file names to merge